Designed for non-coding RNA
CRISPRlnc:database and predict tool
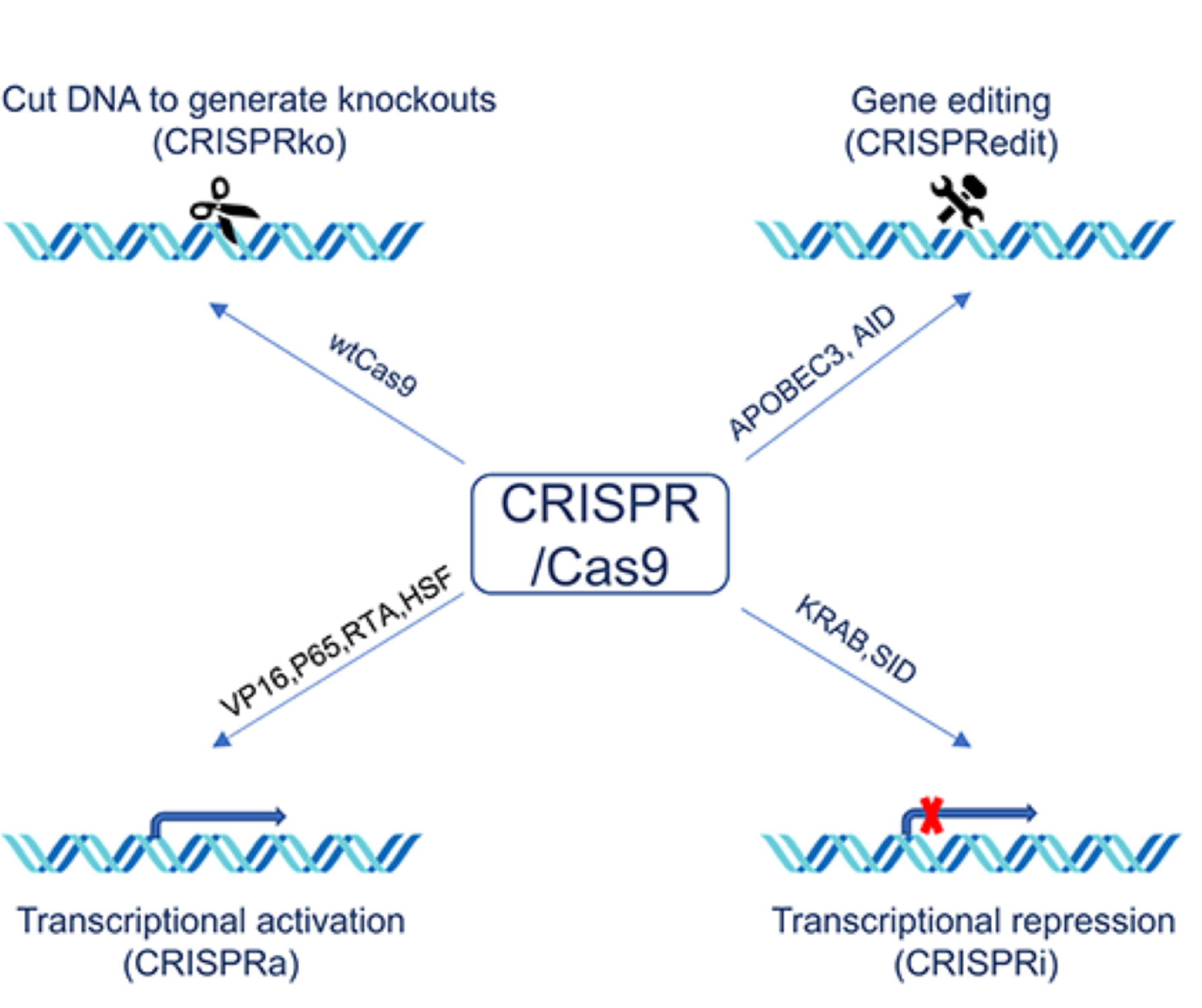
CRISPR-Cas9 is a gene editing technique that performs specific DNA modifications on targeted genes. The simplicity and efficiency of this technology also make it the most widely used method in gene editing.
In addition to the widespread use of CRISPR-Cas9 for coding genes, more and more research teams are now applying it for editing non-coding genes.
One-stop collection and sorting of CRISPR-CAS9 sgRNA data designed for non-coding RNA, and launch of the exclusive CRISPR-CAS9 sgRNA design tool for non-coding RNA.

Database
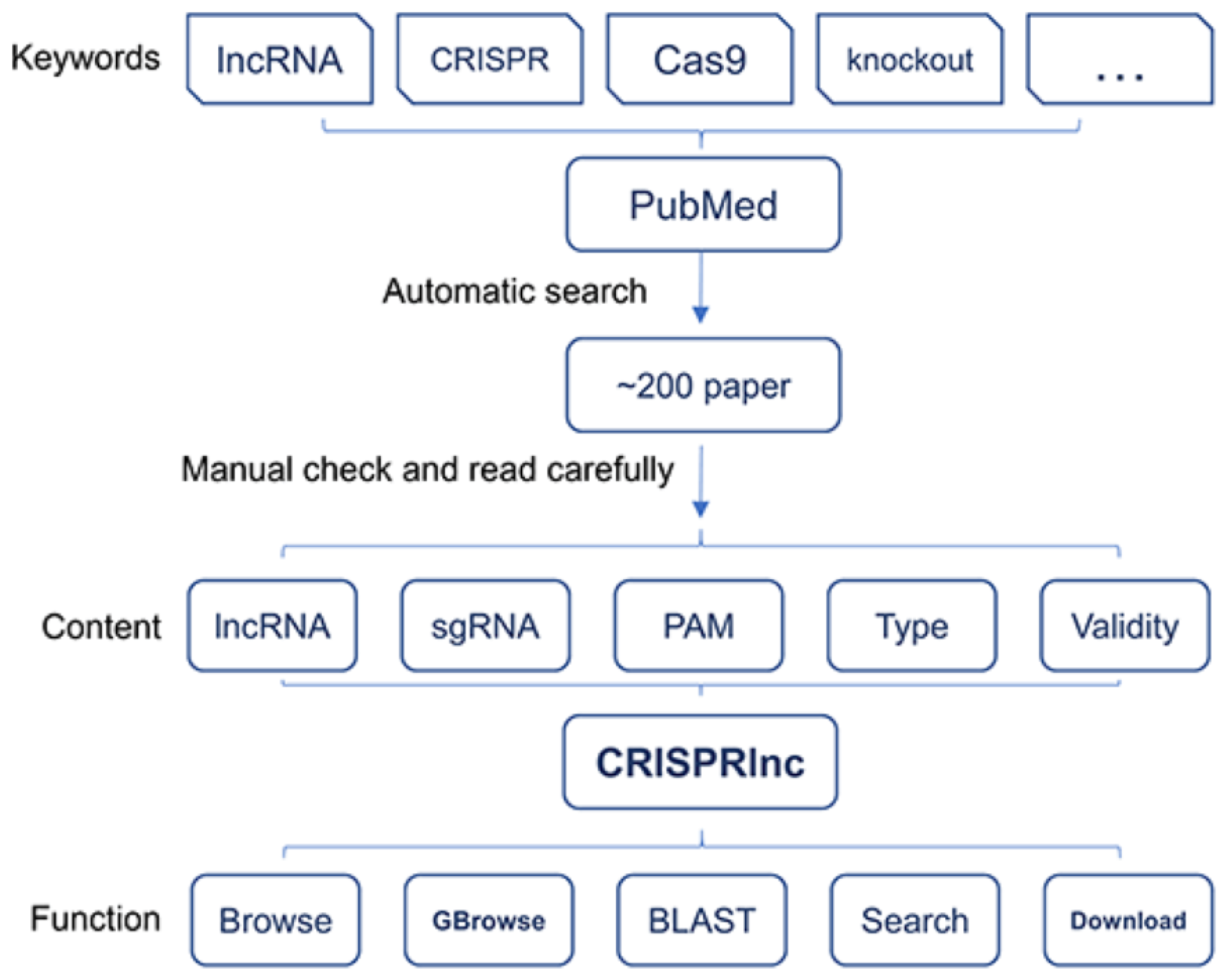
CRISPRlnc database is a manually curated database of validated CRISPR/Cas9 sgRNAs for lncRNAs from all species. After manually reviewing more than 200 published literature, CRISPRlnc collected hundreds of lncRNAs and thousands of validated sgRNAs. We handled the ID, position in the genome, sequence and functional description of these lncRNAs, as well as the sequence, PAM motif, CRISPR type and validity of these sgRNAs.
Until now, tools with various characteristics for ab initio designing sgRNA are crowded, whereas systematic collection of validated sgRNA is rare. As the first database against the validated sgRNAs of lncRNAs, CRISPRlnc will provide a new and powerful approach for genome editing of lncRNAs. We believed that our CRISPRlnc database will thrive in the fields related to lncRNA and CRISPR/Cas9 studies, and will be popular among people in these fields.
Predict Tool
The efficiency of CRISPR-Cas9 gene editing largely depends on the selection of sgRNA, so it is crucial to design an efficient sgRNA.
We evaluated the performance of a series of guide RNA design tools in the CRISPR-Cas9 system under both coding and non-coding datasets. And this efficient sgRNA prediction tool is designed for the preference of sequence, GC content, secondary structure and other characteristics for the two mechanisms of non-coding genes.